Nir Research Group
DNA Nanomachine Lab
Ben Gurion University of the Negev
DNA Nanomachines
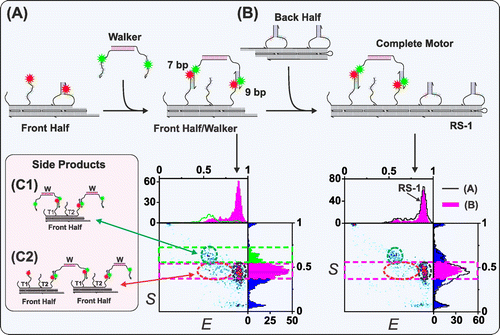
Autonomous motor: We constructed a bipedal autonomous DNA-motor with a coordinate activity between the two motor legs and monitored its activity using SMF techniques. The measurements are done in-situ which enables monitoring the motor’s progress and structural dynamics without disturbing its activity. Our kinetic measurements of the motor’s assembly and activity indicate that it takes dozens of seconds to complete reactions, rather than hours, if components are properly designed. However, we monitor side reactions that significantly reduce the yield of the reaction and resulting in defected motors. We are now working on implementing new strategies for motors’ preparation which will prevent side reactions, altogether, resulting in much higher yield. On the methodological side, we have measured the motor’s dynamics and its interaction with its energy source, a DNA-fuel, in equilibrium and non-equilibrium conditions. Our work demonstrates that by using SMF, one can construct a DNA-machine and monitor its activity in ways not possible with conventional methods. We demonstrate that our methods enable simultaneous in-situ monitoring of the motors efficiency, integrity and activity.
Non-autonomous motor: We constructed a two-leg motor which walks on a DNA-origami track and use energy in the form of fuel/anti-fuel ss-DNAs. The motor successfully completes more than 15 steps and we are now working on increasing the stepping efficiency, speed and walking range. Kinetic measurements indicate that it is possible to significantly improve this kind of motors, such that they will have far higher efficiency and speed. The motor will be capable of maneuvering molecules of interest, e.g. nanoparticles, to a specific location and orientation. Later on, we will use these motors in a device which will maneuver nano-particles in respect to each other in electro-optic device, and a device that exert force on foreign molecules.

Methodological Development
Our group specializes in developing SMF spectroscopical techniques. We are currently working on several methodological developments which will significantly improve the SMF resolution, accuracy and stability.
Photons distribution analysis (PDA): In a single-molecule fluorescence measurement of freely diffusing samples, photons belonging to a single-molecule (‘burst’) are analyzed in terms of their number and not distribution. As a result, static and dynamic heterogeneity are not distinguishable, and often, data is wrongly interpreted. We developed a mathematical algorithm that analyzes the photon distribution inside the burst, indicating whether a single-molecule event undergoes dynamics or it is purely static. Using our method, it is now possible to analyze complicated dynamical behavior in ways not possible with the conventional method. We believe our straightforward, robust and informative approach will be adapted and widely used for analyzing SMF data.
Photon-by-Photon Hidden Markov Model: We develop several methods to measure reaction kinetic profiles at the single-molecule level and successfully implement them to monitor DNA-motors assembly and activity reactions. we introduce H2MM, a maximum likelihood estimation algorithm for photon-byphoton analysis of single-molecule fluorescence resonance energy transfer (FRET) experiments. H2 MM is based on analytical estimators for model parameters, derived using the Baum−Welch algorithm. An efficient and effective method for the calculation of these estimators is introduced. H2 MM is shown to accurately retrieve the reaction times from ∼1 s to ∼10 μs and even faster when applied to simulations of freely diffusing molecules.


Microfluidics based DNA Machines
Realization of bioinspired molecular machines that can perform many and diverse operations in response to external chemical commands is a major goal in nanotechnology, but molecular machines respond to only a few sequential commands in general. Lack of effective methods for introduction and removal of command compounds and low efficiencies of the reactions involved are major reasons for the limited performance. We introduce here a user interface based on a microfluidics device and single-molecule fluorescence spectroscopy that allows efficient introduction and removal of chemical commands and enables detailed study of the reaction mechanisms involved in the operation of synthetic molecular machines.
In the motor operated without strand removal, redundant strands interfere with motor operation and reduce its performance. The microfluidics also enabled computer control of motor direction and speed.
Our universal interface enables two-way communication between user and molecular machine and, relying on concepts similar to that of solid-phase synthesis, removes limitations on the number of external stimuli. This interface, therefore, is an important step toward realization of reliable, processive, reproducible, and useful externally controlled DNA nanomachines.


Fabrication of DNA origami structures
Organizing DNA origami building blocks into higher order structures is essential for fabrication of large structurally and functionally diverse devices and molecular machines. Unfortunately, the yields of origami building block attachment reactions are typically not sufficient to allow programed assembly of DNA devices made from more than a few origami building blocks. Efficient fabrication of structurally and functionally diverse nanomolecular devices and machines by organizing separately prepared DNA origami building blocks into a larger structure is limited by origami attachment yields.



